 Non-B DNA Predictor
Non-B DNA Predictor
An application for identifying intramolecular non-B DNA motifs
Non-B DNAs
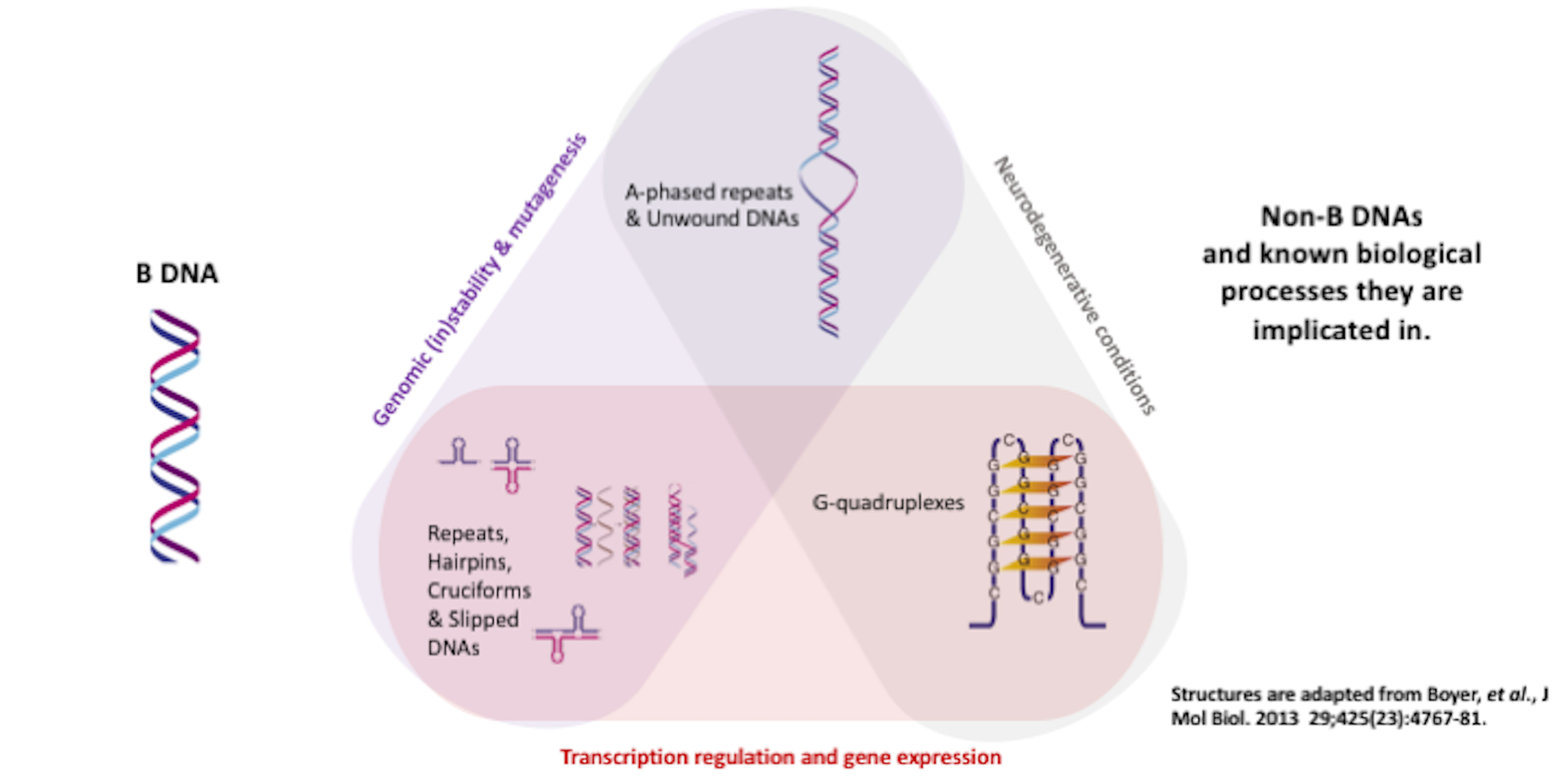
DNA was first identified in the late 1860s by Swiss chemist Friedrich Miescher. In the decades following, other researchers revealed additional details about the DNA molecule, including its primary chemical components and the ways in which they joined with one another. These laid the foundation for the ground breaking description of the DNA by Watson and Crick in 1953. It is now known that genomic DNA is not limited to the confines of the canonical B-forming DNA duplex described by Watson and Crick, but includes other different types of secondary structures collectively termed non-B DNA structures. Non-B DNA have recently garnered much interest because of their important roles in a variety of biological processes such as DNA replication, telomere maintenance, gene expression, viral replication, immune evasion, neurological disorders and cancers.
Non-B DNA Predictor
Non-B DNA Predictor identifies non-B DNA motifs in DNA (and RNA where applicable) sequences. Non-B DNA Predictor is based on a backend R package gquad. This app is a valuable analytical tool for experimental biologists with little computational skills. The motifs that can be identified are A-phased DNA repeats, G-quadruplexes (with or without overlaps), intramolecular triplexes (with or without overlaps), slipped motifs, short tandem repeats, triplex forming oligonucleotides and Z-DNA motifs.
To carry out predictions, the user can either paste raw nucleotide sequences or upload a fasta file or provide GenBank accession numbers into the appropriate pane. Results providing information on the positions, sequence compositions and lengths of predicted motifs are generated and can be downloaded as a csv file. If more than one sequence is provided, an input ID is generated for motifs predicted from each input sequence. For G-quadruplexes, slipped motifs and Z-DNA motifs, the degree of likeliness for a predicted motif to be formed is computed and scored as ** (more likely) or as * (less likely). Users are advised to limit each run to a maximum of 200KB of data. For runs exceeding 200KB, users may contemplate gquad. Please note that results are not stored on our server. Should you have comments, questions or suggestions, contact the maintainer (hajoge[at]uwo.ca).
Sample sequences
Sample sequences are provided for demonstration purpose. The sequences are in three formats and has been put together in a folder (total size of 184KB), which can be downloaded from here as a zip file (57KB). "Sample.fasta" should be uploaded through the "FASTA File" tab, while "SampleGenBank.txt" and "Sample.txt" should be copied into the "GenBank Number" and the "Nucleic Acid Sequences" tab areas, respectively. Each sample file includes (1) a cloned human sequence, (2) human epidermal growth factor receptor, (3) human transketolase like 1 gene and (4) human vascular endothelial growth factor A.
A-phased DNA repeats
A-phased DNAs are right-handed helical structures like B-DNAs, but with difference in geometric attributes. They are found near origins of replications and transcription factor binding sites; and have been implicated in transcription stalling.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download
G Quadruplex Motifs not including Overlaps
G-quadruplexes (G4s) are formed in guanine rich nucleic acids. G4s are abundant in telomeres and they have been implicated in DNA replication, gene expression and genome stability. G4s are also potential antiviral targets.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download
G Quadruplex Motifs including Overlaps
G-quadruplexes (G4s) are formed in guanine rich nucleic acids. G4s are abundant in telomeres and they have been implicated in DNA replication, gene expression and genome stability. G4s are also potential antiviral targets.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download
Intramolecular triplexes (H-DNA) Motifs not including Overlaps
H-DNAs are intramolecular three-stranded hydrogen-bond nucleic acids. H-DNAs are implicated in mutagenic and DNA repair activities.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download
Intramolecular triplexes (H-DNA) Motifs including Overlaps
H-DNAs are intramolecular three-stranded hydrogen-bond nucleic acids. H-DNAs are implicated in mutagenic and DNA repair activities.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download
Slipped motifs
Slipped-DNAs result from mispairing of repeat units on complementary strands. The presence of these structures are implicated in several neurodegenerative and neuromuscular diseases.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download
Short tandem repeats
Short tandem repeats (STRs) are present in 3% of the human genome and mostly found in non-coding regions. They have been implicated in transcription regulation, gene expression and in certain neurodegenerative diseases.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download
Triplex forming oligonucleotides
Triplex forming oligonucleotides are capable of forming both intramolecular and intermolecular three-stranded structures, which can be highly mutagenic.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download
Z-DNA motifs
Z-DNA is a left-handed helical highly stable DNA structure. Z-DNAs have been linked to the induction of genomic instability in mammalian and bacterial cells.
Input
Please enter raw(plain) nucleotide sequence(s), separated by commas.
DO NOT enter FASTA here, upload fasta as a file in the right tab!
Please enter valid GenBank accession number(s), separated by commas.
Example of two valid accession numbers are provided in the pane. Click "Run" to test the example, or replace with your valid numbers and click "Run".
This app retrieves sequence(s) from National Center for Biotechnology Information (NCBI).
If your data size is within upload limit of 200KB and an error occurs, try again later when connection with NCBI may be better!
Please upload a valid FASTA formatted file
Prediction Results
Download
Download